- 1Department of Biological Sciences, University of Alabama, Tuscaloosa, AL, United States
- 2Department of Chemical and Biological Engineering, University of Alabama, Tuscaloosa, AL, United States
- 3Center for Convergent Biosciences and Medicine, University of Alabama, Tuscaloosa, AL, United States
- 4Alabama Life Research Institute, University of Alabama, Tuscaloosa, AL, United States
- 5Department of Microbiology, Heersink School of Medicine, University of Alabama at Birmingham, Birmingham, AL, United States
- 6Department of Biological Sciences, University of Texas at Dallas, Richardson, TX, United States
BRCA1/2 genes are considered tumor suppressor genes and help repair damaged DNA. Pathogenic germline mutations of BRCA1/2 genes are the most common hereditary cause of breast cancer and ovarian cancer. It has been established that BRCA1 mutations increase the risk of brain metastasis compared to the BRCA1 wildtype, and once metastasis occurs to the brain the disease is considered uncurable. The blood-brain barrier (BBB) is essential for maintaining and regulating homeostasis of the central nervous system and is composed of highly specialized brain endothelial cells. Using a human induced pluripotent stem cell (hiPSC) based model, we characterized an hiPSC line from an invasive cancer patient harboring a BRCA1 mutation. This patient-derived hiPSC line can be utilized to study BBB properties as after differentiation into brain-like endothelial cells (BECs), BECs derived from this line express BBB markers such as tight junction proteins, and functional efflux transporters. Future application of patient-derived stem cell models could provide a platform to discover genetic predispositions to BBB disruption in individuals with BRCA1 mutations, as well as the potential molecular mechanisms contributing to brain metastasis.
Introduction
In the United States, cancer continues to be the second-leading cause of death after cardiovascular disease (Siegel et al., 2024; Wang et al., 2016; Nagai and Kim, 2017). BReast CAncer (BRCA) genes, including BRCA1 and BRCA2, are tumor suppressor genes, whose role is established to repair damaged DNA (Boulton, 2006; Mai et al., 2009). Consequently, mutations in BRCA1/2 genes hinder proper DNA repair and have been shown to increase the malignancy risk for hereditary cancers, especially breast and ovarian cancers (Gudmundsdottir and Ashworth, 2006; Ratner et al., 2019). BRCA1 mutations have also been associated with earlier diagnosis and progression of brain metastases (Ratner et al., 2019). In ovarian cancer patients, the BRCA1 mutation has shown a 4-fold greater risk for brain metastases compared to the BRCA1 wildtype and has led to an 8-month earlier diagnosis of brain metastasis (Ratner et al., 2019). Similarly, for central nervous system metastatic breast cancer, BRCA1/2 mutations have shown a younger median age for diagnosis, a greater incidence of high-grade tumors, and a lower survival time post-diagnosis than wild-type BRCA1 individuals (Ben-Zion Berliner et al., 2024). To metastasize to the brain, cancer cells in circulation must interact with and penetrate the blood-brain barrier (BBB) (Arshad et al., 2011).
The BBB is comprised of highly specialized brain endothelial cells (BECs) that encompass the cerebral vasculature (Abbott et al., 2010; Knopp et al., 2022). The BBB is essential for the maintenance and regulation of the neural microenvironment and prevents the entry of pathogens and toxins into the CNS (Helms et al., 2016; Patabendige and Janigro, 2023). BECs serve as the interface between the circulation and the CNS and express complex tight junctions (Rubin and Staddon, 1999). Due to the presence of efflux transporters and low endocytosis rates, the BBB restricts 100% of large molecule drugs and 98% of small molecule drugs which poses an obstacle for drug delivery of chemotherapeutics (Pardridge, 2007; Hersh et al., 2016).
BBB models present unique challenges as robust in vivo modeling may have interspecies variation, and in vitro modeling of primary cells are limited by passage number and often lose BBB-defining phenotypes when removed from the neural microenvironment (Helms et al., 2016; Obermeier et al., 2013; Andjelkovic et al., 2020; He et al., 2014). Human stem cell (hSC) technology has allowed for the advancement of in vitro modeling and overcomes challenges such as the loss of BBB-defining phenotypes, scalability, and interspecies variations (Obermeier et al., 2013; Verscheijden et al., 2021; Wilhelm and Krizbai, 2014).
Previous studies have successfully used patient derived stem cells to model specific disease contexts in the BBB (Vatine et al., 2017; Yucer et al., 2021; Ozgür et al., 2023). Patient derived hiPSCs harboring MCT8 mutations have been used to model the BBB and study drug transport in a diseased BBB (Vatine et al., 2017).
It has also been demonstrated that hiPSC-BECs derived from Huntington’s disease patients had key transcriptional and functional differences compared to healthy control hiPSC-BECs, indicating a crucial deficit in BBB function in this neurodegenerative disease context (Lim et al., 2017). BRCA1 mutated cells lines have been used to model fallopian tube epithelium and recapitulated ovarian carcinogenesis (Yucer et al., 2021). Patient derived models provide the ability to model human diseases with a specific genetic predisposition (Vatine et al., 2017). However, to our knowledge, hiPSC-BEC models derived from BRCA1 mutated patients have not been reported.
In this study, we use an induced pluripotent stem cell model, where over the period of 13 days, patient-derived hiPSCs are differentiated, expanded, and purified with the result of patient-derived brain-like endothelial cells (Lippmann et al., 2012; Espinal et al., 2022; Lippmann et al., 2014). Recent work has demonstrated that while BEC differentiation is robust, some human iPSC lines may differ in the BBB properties exhibited after differentiation (Vatine et al., 2017; Ozgür et al., 2023; Lim et al., 2017; Patel et al., 2017). For example, the SBAD0201 stem cell line showed non-functional efflux transporter P-glycoprotein (P-gp) (Ozgür et al., 2023). Throughout the study we use the well-characterized hiPSC line IMR90-4 for comparison of key BBB marker expression after differentiation. The goal of this study was to characterize the BEC differentiation and resulting BBB-like properties of the BRCA1 mutated patient-derived stem cell line CS79iBRCA-n2 in vitro.
Methods
Cell lines
The CS79iBRCA-n2 cell line was purchased from Cedars-Sinai Biomanufacturing Center. Lymphoblastic cells were first harvested from a female early-onset, stage IIIC ovarian cancer patient with a BRCA1 mutation (IVS5+1G>A, located at the junction between exon five and intron 6). The CS79iBRCA-n2 cell line was reprogrammed from the Cedars-Sinai Biomanufacturing Center parent cell line 2012-0872 4079. Lymphoblastic cells were reprogrammed to iPSCs via an engineered episomal plasmid containing reprogramming factors Oct3/4, Sox2, KLF4, L-Myc, shp53, and Lin28. The IMR90-4 cell line was purchased from WiCell (WISCi004-B) where it was reprogrammed using viral transfection methods from a female parent cell line, IMR90.
Culturing of hiPSCs
hiPSCs were cultured on Matrigel (Corning; 354234) coated 6-well plates and maintained in StemFlex Medium with supplement (Gibco; A3349401), changed daily (Lippmann et al., 2012; Espinal et al., 2022; Lippmann et al., 2014; Stebbins et al., 2016; Gomes et al., 2019). When iPSCs reach 70% confluence, cells were dissociated from the well using Versene (Gibco; 15040066) and passed at 1:3, 1:6, and 1:12 split ratios onto Matrigel coated 6-well plates. Cells were continuously maintained in StemFlex medium, changed daily. Passages 15–25 were used for differentiations.
Differentiation of hiPSCs into brain-like endothelial cells
When iPSCs reach 80% confluence, cells were differentiated according to the protocol we and others have used previously (Lippmann et al., 2012; Espinal et al., 2022; Stebbins et al., 2016). Cells were seeded onto Matrigel-coated T75 flasks (VWR; 10861-646) at 10,000 cell/cm2 and maintained in StemFlex Medium with supplement for 3 days, with media changed daily. Differentiation into induced brain endothelial cells (BECs) was initiated after 3 days with the addition of unconditioned medium (UM) for 6 days, changed daily (Stebbins et al., 2016). UM media is composed of 89% DMEM/F12 (ThermoFisher; 11330-032), 10% Knockout serum replacement (KOSR) (Gibco; 10828028), 1% Non-essential amino acids (NEAA) (ThermoFisher; 11140-050), 0.5% Glutamax (ThermoFisher; 35050061), and 0.005% β-mercaptoethanol (β-ME). Differentiation was continued with the use of endothelial cell (EC) medium, containing hESFM (ThermoFisher; 11111-044) supplemented with 1% B27 (Gibco; 17504044), 10 µM retinoic acid (RA) (Sigma; R2625), and 20 ng/mL basic fibroblast growth factor (bFGF) (PeproTech; 100-18B-250UG), for 2 days, with media changed daily.
On Day 8, cells are dissociated and singularized using Accutase (StemCell Technologies; MSPP-07920). BECs were then purified by seeding onto ECM coated plates which selectively isolates cells expressing BEC phenotypes and not neural cells (Sigma; C5533-5MG, F1141-5MG). Cells were replated at a seeding density of 500,000 cells/well for 24-well plates and 1 million cells/well for 12-transwell plates in EC + RA medium. Media was changed to EC medium without bFGF and RA supplements (EC - -) 1 day later. BECs were validated through measuring trans-endothelial electrical resistance (TEER) on Day 9 and Day 10, through immunostaining for BEC markers on Day 10, and through functional assays on Day 10.
P-gp functional assay
Using previously published protocols, P-glycoprotein (P-gp) function in hiPSCs and BECs was determined through the measurement of the accumulation of the P-gp substrate, Rhodamine 123 (R123) (Lippmann et al., 2012; Stebbins et al., 2016; Kim et al., 2019). The potent P-gp inhibitor, Valspodar (Sigma; SML0572-5MG), was utilized to assess P-gp activity for comparison. The cells were washed once with warmed Hank’s Buffer Saline Solution (HBSS) (ThermoFisher; 14065056) and pretreated with or without 10 µM of the inhibitor, Valspodar, with HBSS for 1 h at 37°C and 5% CO2. After pretreatment, the cells were then incubated with 10 µM of R123 with or without 10 µM of Valspodar in HBSS for 2 h in 37°C and 5% CO2. After incubation, the cells were washed with cold PBS twice and 200 µL of Radioimmunoprecipitation assay (RIPA) buffer was applied. After RIPA buffer application, the plate was then placed on a rotator to shake for 10 min at room temperature while protected from light. The plate fluorescence was then quantified on a plate reader (Molecular Devices SpectramaxiD3). BCA assays (ThermoFisher; 23227) were used to normalize the fluorescent values to account for the number of cells present.
BCRP functional assay
Breast Cancer Resistance Protein (BCRP) activity in hiPSCs and BECs was measured through the accumulation of the fluorescent BCRP substrate, Hoechst 33342 (ThermoFisher; 62249) (Lippmann et al., 2012; Stebbins et al., 2016). BCRP inhibitor, Ko143 (Enzo: 89158-270), was used as comparison (Kim et al., 2019; Allen et al., 2002; Paturi et al., 2010). The cells were washed once with warm Hank’s Buffer Saline Solution (HBSS) and pretreated with or without Ko143 at 1 µM concentration in HBSS for 1 h at 37°C and 5% CO2. After pretreatment, the cells were then incubated with 10 µM of Hoechst 33342 in HBSS with or without inhibitor for 2 h at 37°C and 5% CO2. The cells are then washed twice with 500 µL of cold PBS, and then 200 µL of RIPA buffer was applied. The cells were then placed on a rotator for 10 min protected from light. Fluorescence was measured on a plate reader (Molecular Devices SpectramaxiD3). To normalize the fluorescent values, BCA assays (ThermoFisher; 23227) were performed, and OD values were used to normalize.
RNA isolation and quantitative PCR
RNA was isolated using the NuceloSpin RNA kit (Macherey-Nagel; 740955.50) and cDNA was prepared using LunaScript RT (New England BioLabs; E3010). SYBR green qPCR was run for human POU5F1 (Oct4) forward primer 5′–CCCCAGGGCCCCATTTTGGTACC – 3′ and reverse primer 5’ – ACCTCAGTTTGAATGCATGGGAGAGC – 3’. 18S rRNA was used as a reference gene for normalization, forward primer 5′–GTAACCCGTTGAACCCCATT – 3′ and reverse primer 5′–CCATCCAATCGGTAGTAGCG – 3′. qPCR was run on a QuantStudio 3 Real-Time PCR system (ThermoFisher). Relative gene expression was calculated using the ΔΔCt method.
Immunostaining
Both CS79-derived and IMR90-4 derived BECs were immunostained following day 10 of differentiation (Stebbins et al., 2016). CS79-derived hiPSCs were immunostained at 3 days post passaging. Cells were fixed depending on the antibody with either ice cold methanol or 4% paraformaldehyde (diluted in PBS) for 15 min and blocked in 10% Fetal Bovine Serum (FBS) (Table 1) (Stebbins et al., 2016). Markers for immunostaining include Claudin-5, Occludin, ZO-1, P-gp, GLUT1, VE-Cadherin, PECAM-1, and BCRP. Expression was visualized using a Nikon Ti2 inverted epifluorescence microscope equipped with a Qi2 camera (Nikon, Tokyo, Japan) using NiS Elements software version AR.5.30.05 for acquisition. Images were analyzed using ImageJ Software (FIJI).
Western blot
The protein amounts were derived from BCA assays (ThermoFisher; 23227). The protein samples were heated for 5 min at 95°C before being loaded onto Bolt 4%–12% Bis-Tris Plus protein gels (ThermoFisher; NW04120BOX, NW04125BOX) along with Peacock Plus Prestained Protein Marker (Biotium; 21531) and transferred to nitrocellulose membranes. In the case of BCRP and GLUT1 targeting, 10% β-Mercaptoethanol (β-ME) instead of heat was added to the protein samples. Following 1 h blocking at room temperature with tri-buffered saline +0.1% Tween 20 (1x TBST) and 5% nonfat dry milk, the respective primary antibodies were used for targeting and remained on the membrane overnight at 4°C (Table 1). The next day the blots were washed in TBST (3x) and given goat anti-mouse IgG (H + L) Alexa Fluor 488 conjugate secondary antibody (ThermoFisher; A-11001) for 1 h at room temperature while shaking. For P-gp, goat anti-rabbit IgG (H + L) Alexa Fluor 488 conjugate second antibody (ThermoFisher; A-11034) was used instead. After washing in TBST (3x), the blots were imaged on an iBright FL1500 Imaging System instrument (ThermoFisher) with SuperSignal West Pico PLUS Chemiluminescent Substrate (ThermoFisher; 34577) for visualization.
TEER measurement
On day 8, purified BECs were seeded onto ECM coated 12 well transwell (Corning; 3460). TEER was measured using a EVOM II instrument (World Precisions). For an 8-day duration, the basal and apical layers were probed for TEER and received changes in EC- - media. A media-only transwell was used for temperature adjustment before the probe handled any measurements. TEER was additionally measured on three independent passages of CS79-hiPSCs for a period of 5 days post expansion.
Dextran uptake assay
On day 8, purified CS79-derived BECs were seeded onto ECM coated 24 well plates at 500,000 cells/cm2. CS79-hiPSCs were seeded onto the same plate at the same density. On day 10, cells were treated with 10 kD fluorescein dextran at 1 mg/mL (ThermoFisher; D1821). Plates were either incubated at 37°C + 5% CO2 or at 4°C for 30 min. Cells were then removed from incubation and lysed using 500 μL RIPA buffer per well. Fluorescence values were measured using a plate reader (Molecular Devices SpectramaxiD3). A BCA assay was then performed, and OD values were used for normalization.
Sodium fluorescein transport
We performed sodium fluorescein permeability assays as previously described (Stebbins et al., 2016). Briefly, we resuspend sodium fluorescein (Sigma; F6377-100G) to a stock concentration of 10 mM in PBS and stored it at 4°C protected from light. To make a working concentration, we diluted the stock solution in EC medium for a final concentration of 10 μM. On day 8, cells were seeded at a density of 1 million cells/cm2 onto ECM coated transwells. A blank ECM coated transwell was included to use in later calculations. On day 10, TEER was measured. Once TEER was measured, the medium was removed and replaced with pre-warmed EC medium by adding 500 μL to the top chamber and 1,500 μL to the bottom chamber. The plate was then incubated at 37°C for 60 min. After incubation, TEER was measured again to determine initial barrier integrity. After, medium was aspirated in the top of the transwell and replaced with 500 μL of the 10 μM working concentration of sodium fluorescein in blank and seeded transwells. After 15 min, 150 μL was collected from the bottom chamber of each transwell and placed into a 96-well plate. The removed medium was replaced with 150 μL of pre-warmed EC medium. This process was repeated every 15 min at 30, 45, and 60 min. At 60 min, an additional 150 μL from the top of the transwells was collected and transferred to a 96-well plate in addition to 150 μL of EC medium without sodium fluorescein. The solution from the top chambers were diluted to combat oversaturated signal. The fluorescence values were read using a plate reader (Molecular Devices SpectramaxiD3) at 485 nm excitation and 530 nm emission. We then calculated corrected signal, clearance volumes, the linear slope of clearance volume versus time, and the sodium fluorescein permeability using previously described formulas (Stebbins et al., 2016).
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 9.1.2 unless otherwise stated. Student’s t-test was used to determine significance for pair-wise comparison. Statistical significance was determined for a p-value of 0.05 or less. Error bars represent standard deviation (SD).
Data availability
All data generated or analyzed during this study are included in this published article. hiPSC line CS79iBRCA-n2 available from Cedars-Sinai Biomanufacturing Center iPSC Core Repository.
Results
CS79iBRCA-n2 derived BECs possess endothelial properties
The CS79-hiPSCs were harvested from a BRCA1 mutated invasive cancer patient (Figure 1A) and were then differentiated over the course of 13 days into BECs (Figure 1B). Using a previously published differentiation protocol, we were able to differentiate the CS79-hiPSCs into CS79-derived BECs by seeding the CS79-hiPSCs first onto Matrigel-coated flasks (Espinal et al., 2022; Stebbins et al., 2016). The CS79-iPSCs were then grown in unconditioned medium (UM) and differentiated to an endothelial cell and neural progenitor cell culture mixture (Lippmann et al., 2012; Lippmann et al., 2014). The switch to endothelial cell (EC) medium allows endothelial cells to be selectively expanded (Lippmann et al., 2014). On day 8, the BECs were purified onto extracellular matrix (ECM) coated plates (collagen IV, fibronectin, molecular water) which resulted in a purified monolayer of BECs (Figure 1B). We performed qPCR on CS79iBRCA-n2 hiPSCs and CS79-derived BECs to look at pluripotency marker Oct4 (POU5F1) where we saw a decrease of expression after differentiation (Figure 1C). To determine the barrier properties of the differentiated BECs, trans-endothelial electrical resistance (TEER) readings were obtained beginning on day 8 of the differentiation, when the BECs were seeded onto ECM coated plates (Stebbins et al., 2016; Sun et al., 2022). TEER was measured daily for 8 days after purification. This was repeated for four independent differentiations of CS79-derived BECs and results were similar throughout where TEER remained high until 5 days post purification when TEER began to decrease (Figure 1D). TEER was also recorded for three independent passages of CS79iBRCA-n2 hiPSCs to establish a baseline value (Figure 1D). Additionally, for a control cell line we used a well characterized hiPSC cell line, IMR90-4, and repeated the differentiation protocol. We then recorded TEER of three independent differentiations for 8 days post purification (Figure 1E).
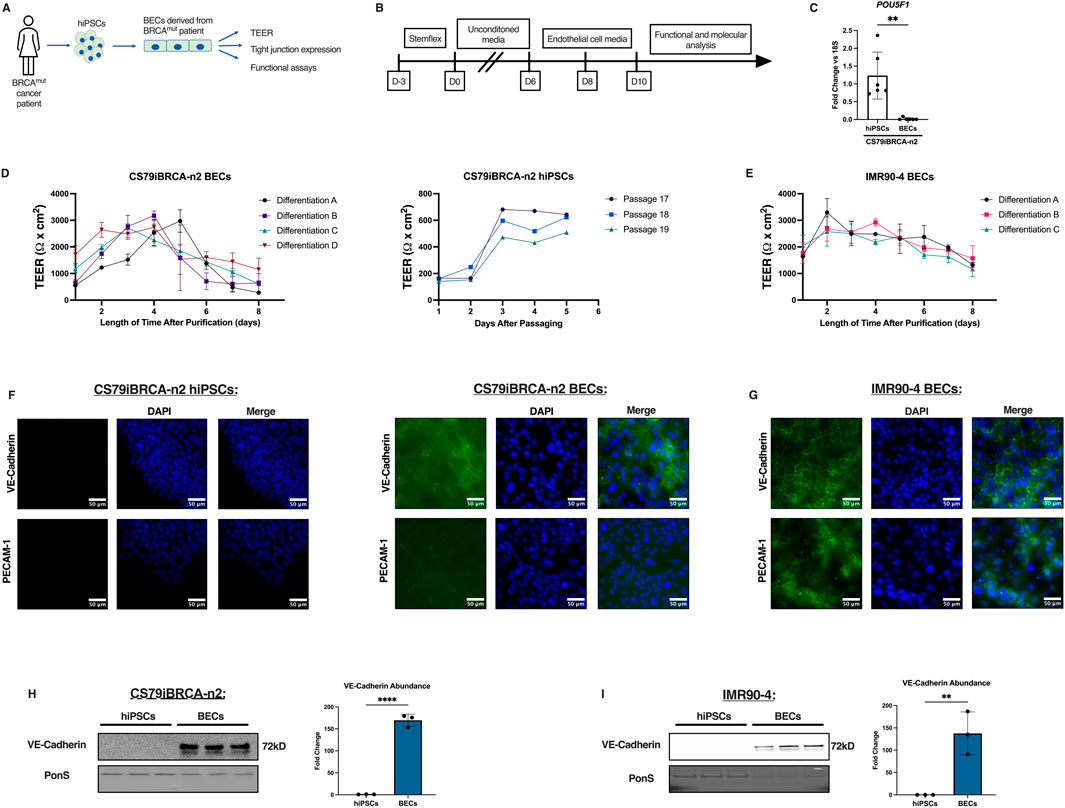
Figure 1. CS79iBRCA-n2-derived BECs have endothelial properties. (A) Graphic outlining the process of harvesting the hiPSCs and their differentiation into BECs. (B) Timeline outlining the differentiation process. (C) qPCR showing expression of pluripotency marker, POUF51, in CS79-hiPSCs compared to CS79-derived BECs (N = 6). (D) TEER measurements from CS79-derived BECs over a period of 8 days post purification from four independent differentiations conducted in triplicate per differentiation (n = 12). TEER measurements of three CS79-hiPSCs passages over a period of 5 days post passaging. (E) TEER measurements from IMR90-4 derived BECs over a period of 8 days post purification for three independent differentiations (n = 9). (F) Representative immunofluorescence images of CS79-hiPSCs and BECs for endothelial markers VE-Cadherin and PECAM-1 (green) with nuclei stained with DAPI (blue). (G) Representative immunofluorescence images of IMR90-4 derived BECs for endothelial markers VE-Cadherin and PECAM-1 (green) with nuclei stained with DAPI (blue). (H) Western blot analysis of CS79-hiPSCs and CS79-derived BECs probing for endothelial marker VE-Cadherin with densitometry quantification. PonS stain shown to show relative protein concentrations loaded for each lane (n = 3). (I) Western blot analyses of IMR90-4 hiPSCs and IMR90-4 derived BECs probing for endothelial marker VE-Cadherin with densitometry quantification. PonS stain shown to show relative protein concentrations loaded for each lane (n = 3). Scale bar = 50 μm. Statistical significance calculated by Student’s t-test, **p < 0.01, ****p < 0.0001. Error bars represent SD.
Immunofluorescence images of CS79iBRCA-n2 hiPSCs were obtained for endothelial markers, vascular endothelial-cadherin (VE-Cadherin) and platelet endothelial cell adhesion molecule (PECAM-1) and showed no presence of endothelial markers in the hiPSCs (Figure 1F). However, the differentiated CS79-derived BECs displayed the localization of VE-Cadherin, whereas PECAM-1 showed limited expression and was not localized in the CS79-derived BECs (Figure 1F). Immunofluorescence images of the IMR90-4 BECs showed expression and localization of both VE-Cadherin and PECAM-1 (Figure 1G). To further confirm endothelial properties, Western blot analyses were performed for VE-Cadherin. Signal was observed in the CS79-derived BECs at 72 kDa corresponding to VE-Cadherin and no signal was observed for CS79-hiPSCs (Figure 1H), which was similar to the immunofluorescent staining images. We also performed Western blot analyses for VE-Cadherin on IMR90-4 hiPSCs and BECs, and observed signal in the IMR90-4 BECs and not in the IMR90-4 hiPSCs (Figure 1I). Together these data demonstrate that CS79iBRCA-n2 derived brain-like endothelial cells possess barrier function and some endothelial characteristics.
Tight junction expression of CS79iBRCA-n2 derived BECs
While hiPSCs produce tight junctions, they remain an important aspect of BBB function (Sun et al., 2022; Gastfriend et al., 2018). To characterize how the CS79-hiPSCs and the differentiated CS79-derived BECs expressed tight junction proteins, we measured the expression and localization of Zona Occludens-1 (ZO-1), Occludin, and Claudin-5 (Sun et al., 2021). Immunofluorescence images of the hiPSCs were obtained for ZO-1, Occludin, and Claudin-5. The hiPSCs expressed a high signal intensity for Claudin-5, which should be taken into consideration when utilizing this cell line (Figure 2A). The staining images revealed low signal intensity in the Occludin and ZO-1 tight junction proteins, indicating a low presence in CS79-hiPSCs (Figure 2A). However, when the same markers were measured in the differentiated CS79-derived BECs, we observed an increase in signal intensity in Occludin and ZO-1, suggesting the strong expression and localization of the tight junction proteins (Figure 2A). Additionally, we performed immunostaining on IMR90-4 BECs to show expression and localization of Claudin-5, Occludin, and ZO-1 (Figure 2B). Western blot analyses were conducted on CS79-hiPSCs and BECs using specific antibodies to confirm the presence of the tight junction proteins, ZO-1, Occludin, and Claudin-5. We obtained signal in the BEC samples at 225 kDa (corresponding to ZO-1), 55 kDa (corresponding to Occludin), and 23 kDa (corresponding to Claudin-5) respectively (Figure 2C).
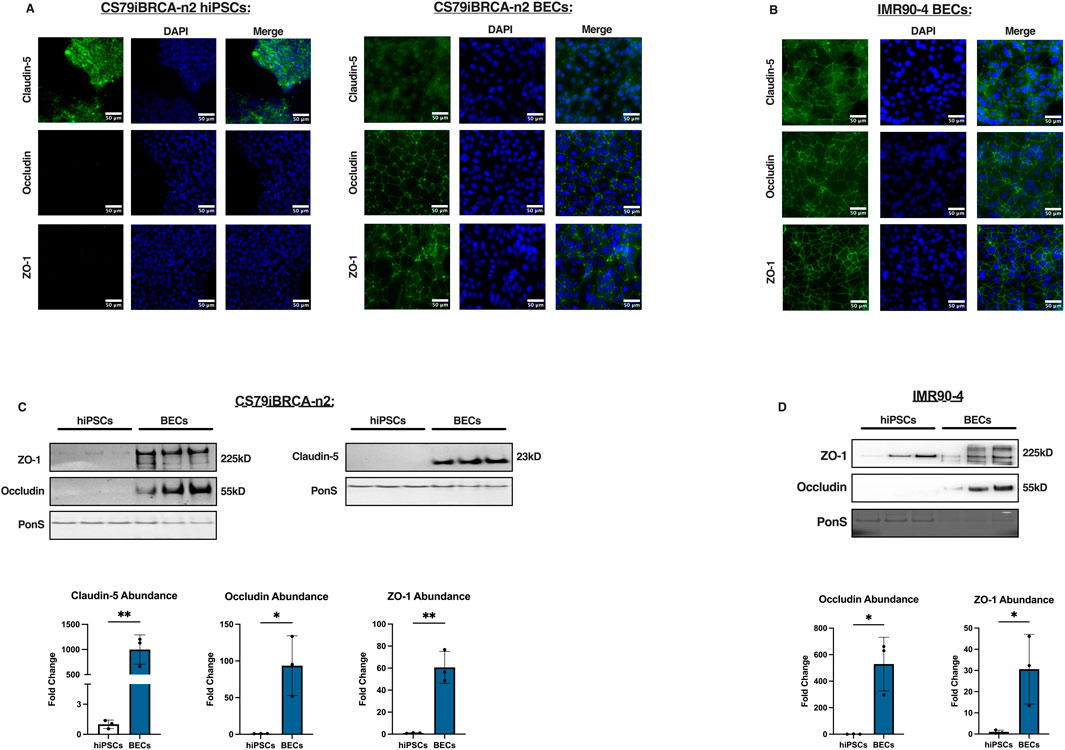
Figure 2. Tight junction expression of CS79iBRCA-n2-derived BECs. (A) Representative immunofluorescence images of CS79-hiPSCs and CS79-derived BECs for the tight junction proteins, ZO-1, Occludin, and Claudin-5 (green) and nuclei stained with DAPI (blue). Scale bar = 50 μm. (B) Representative immunofluorescence images of IMR90-4 derived BECs for tight junction proteins Claudin-5, Occludin, and ZO-1. Scale bar = 50 μm. (C) Western blot analyses of CS79-hiPSCs and CS79-derived BECs probing for tight junction proteins, ZO-1, Occludin, and Claudin-5. PonS stain shown to show relative protein concentrations loaded for each lane (n = 3). Quantification of western blots for ZO-1, Occludin, and Claudin-5 (n = 3). (D) Western blot analyses of IMR90-4 hiPSCs and IMR90-4 derived BECs probing for ZO-1 and Occludin. PonS stain shown to show relative protein concentrations loaded for each lane (n = 3). Quantification of western blots for ZO-1, Occludin, and Claudin-5 (n = 3). Statistical significance calculated by Student’s t-test, *p < 0.05, **p < 0.01. Error bars represent SD.
Results demonstrate that there was a significantly higher abundance as seen by Western blotting of the tight junction proteins, ZO-1, Occludin, and Claudin-5, in the CS79-derived BECs than the hiPSCs (Figure 2C). We additionally performed Western blotting on IMR90-4 hiPSCs and BECs to observe expression of ZO-1 and Occludin which can be looked at as a reference of expression after BEC differentiation (Figure 2D). Together, these data suggest that the BEC differentiation increases the expression of tight junction proteins that contribute to barrier function.
CS79iBRCA-n2 derived BECs express and localize transporter proteins
Next, we sought to observe other BBB properties such as the expression of nutrient transporters and multi-drug efflux transporters (Lippmann et al., 2012). Immunofluorescence images revealed that the nutrient transporter GLUT1 and efflux transporter P-gp was expressed and localized in the CS79-hiPSCs (Figure 3A). Expression of efflux transporter Breast Cancer Resistance Protein (BCRP) was not observed in the CS79-hiPSCs (Figure 3A). Immunofluorescence images of the CS79-derived BECs confirmed the expression and localization of nutrient transporter GLUT1, and the efflux transporters, P-gp and BCRP (Figure 3A). Additionally, expression and localization of GLUT1, BCRP, and P-gp was observed in IMR90-4-derived BECs was observed (Figure 3B). These data suggest that the BEC differentiation process increases some BBB-like transporter expression following hiPSC differentiation.

Figure 3. Characterization of the expression and localization of transporter proteins in CS79iBRCA-n2-derived BECs. (A) Representative immunofluorescence images for CS79-hiPSCs and CS79-derived BECs for nutrient transporter GLUT1, efflux transporter BCRP, and efflux transporter P-gp (green) and nuclei stained with DAPI (blue). Scale bar = 50 μm. (B) Representative immunofluorescence images for IMR90-4 derived BECs expression of GLUT1, BCRP, and P-gp (green) and nuclei staining with DAPI (blue). Scale bar = 50 μm. Substrate accumulation assays determining the function of the efflux transporters (C) BCRP and (D) P-gp for three independent differentiations conducted in triplicate (n = 9). (E) Uptake assay determining large molecule transport of 10 kD Dextran in CS79-hiPSCs and CS79-derived BECs (n = 9). (F) Sodium fluorescein transport assay in CS79-hiPSCs and CS79-derived BECs (n = 9). Student’s t-test was used to determine statistical significance within the same cell conditions, ****p < 0.0001. Error bars represent SD.
CS79iBRCA-n2 derived BECs have a functional BCRP and P-gp efflux transport
To determine whether efflux transporters are functional in the CS79-derived BECs, we utilized and adjusted previously published substrate accumulation assay techniques to quantify P-gp and BCRP function (Stebbins et al., 2016; Kim et al., 2019; Engdahl et al., 2021). When the CS79-derived BECs were treated with the inhibitor, Ko143, we observed a significant increase in the accumulation of the BCRP substrate, Hoechst 33342, within the BECs. Hoechst 33342 accumulation within the cell increased by +50%, demonstrating that BCRP is functional in the BECs (Figure 3C) (Kim et al., 2019; Allen et al., 2002; Paturi et al., 2010). There was no significant increase in Hoechst 33342 accumulation within the CS79-hiPSCs when they were treated with Ko143 (Figure 3C). When the CS79-derived BECs were treated with the P-gp inhibitor, Valspodar, there was a significant increase in the accumulation of the P-gp substrate, Rhodamine 123 (R123). Valspodar treatment resulted in a +50% increase in R123 accumulation in the CS79-derived BECs, demonstrating functional P-gp is present in the CS79-derived BECs (Figure 3D). When CS79-hiPSCs were treated with Valspodar, there was no significant increase in substrate accumulation (Figure 3D) (Mayer et al., 1997). To further examine functional barrier integrity, we performed a large molecule transport assay using 10 kD FITC-Dextran (Figure 3E). We did not see a significant difference in accumulation between CS79-hiPSCs and CS79-derived BECs.
Additionally, we tested small molecule transport using sodium fluorescein where we saw a decrease of small molecule transport in CS79-derived BECs compared to CS79-hiPSCs (Figure 3F). Taken together we find that our differentiation process drives BBB-like phenotypes with the expression of functional efflux transporters BCRP and P-gp when compared to hiPSCs, but may not have robust transport ability of large molecules.
Discussion
Brain metastasis is more prevalent in patients with BRCA1 mutations (Ratner et al., 2019; Ben-Zion Berliner et al., 2024). Unfortunately, once brain metastasis occurs it is no longer considered curable (Narkhede et al., 2017). In vitro iPSC models used to study genetic mutations are limited due to the loss of patient specific genetic characteristics, as well as limited to number of passages before losing key phenotypic properties (Curry et al., 2015). To represent BRCA1 mutations in vitro, a patient derived hiPSC line, CS79iBRCA-n2, was used that maintained the BRCA1 disease mutation and patient’s genetic background. In this study we utilized a previously described differentiation protocol described to characterize the CS79iBRCA-n2 cell line after differentiation into BECs (Lippmann et al., 2012; Lippmann et al., 2014; Gomes et al., 2019). As a control, we utilized a well-characterized hiPSC line, IMR90-4, which has been extensively used to model the BBB, as well has been used with the differentiation protocol utilized in this study (Lippmann et al., 2012; Espinal et al., 2022; Hollmann et al., 2017; Stebbins et al., 2019).
After differentiation, TEER data across four independent differentiations showed tight monolayers with strong barrier integrity. The CS79iBRCA-n2 cell line has elevated TEER values consistent with other hiPSC lines used to generate BECs (Vatine et al., 2017; Lim et al., 2017; Aragón-González et al., 2024). CS79iBRCA-n2 hiPSCs were immunostained for endothelial specific markers VE-Cadherin and PECAM-1. No expression was visualized as expected. CS79iBRCA-n2-derived BECs were also stained for VE-Cadherin and PECAM-1. VE-Cadherin was well localized and distributed in the CS79-derived BECs. Additionally, VE-Cadherin protein abundance was visualized in CS79-derived BECs and not in CS79-hiPSCs which is consistent with the development of endothelial specific markers after differentiation.
To examine tight junction expression, we performed western blots and immunostaining on CS79iBRCA-n2 hiPSCs and CS79iBRCA-n2-derived BECs. We observed ZO-1, Occludin, and Claudin-5 expression and organization in the BECs which contributes to the selective permeability and barrier integrity of the BBB (Figure 2A). We also saw an increase of ZO-1, Occludin, and Claudin-5 protein abundance after differentiation in CS79-derived BECs (Figure 2C) (Urich et al., 2012). In a human immortalized endothelial cell line, CMEC/D3, low expression of Claudin-5 has been observed (Helms et al., 2016). In a different patient derived BBB model using the cell line CTR54F, low immunofluorescence intensity was observed for Claudin-5 and Occludin (Patel et al., 2017). For tight junction expression, the CS79iBRCA-n2 cell line demonstrates strong localization after BEC differentiation and can be used for relevant studies.
Additionally, we show through immunostaining that the CS79iBRCA-n2 derived BECs expressed drug efflux transporters P-gp and BCRP (Figure 3). To determine functionality of BCRP and P-gp we performed substrate accumulation assays, where functionality was observed in the CS79iBRCA-n2-derived BECs for both P-gp and BCRP. Other patient derived BBB models have seen a non-functional P-gp phenotype (Ozgür et al., 2023). Additionally, primary rodent BEC models have shown a downregulation of key BBB proteins such as GLUT-1 and P-gp (Helms et al., 2016). Interestingly, BCRP and P-gp expression was seen localized in the CS79iBRCA-n2-derived BECs. In addition, to investigate large molecule transport in the CS79-derived BECs and CS79-hiPSCs, we utilized dextran uptake assays. Though no significant difference was observed in large molecule accumulation, this could be contributed to the hiPSCs being grown to a full monolayer. Once grown to 100% confluence, the hiPSCs display some barrier properties, as shown by TEER values of the CS79-hiPSCs (Figure 1D). This could contribute to the lack of difference seen between accumulation. For future applications, it could be useful to be tested against a different barrier forming cell. To investigate small molecule transport, the sodium fluorescein assay displayed a decrease in small molecule transport in the CS79-derived BECs compared to the CS79-hiPSCs. Previous studies characterizing other hiPSC cell lines have demonstrated variation in BBB properties among differing cell lines (Vatine et al., 2017; Ozgür et al., 2023; Lim et al., 2017; Aragón-González et al., 2024). Similarly, the CS79-derived BECs express robust BBB phenotypes such as functional BCRP and P-gp and a trending decrease in small molecule transport. Based on these phenotypes, CS79iBRCA-n2 cell line can prove to be useful when studying P-gp or BCRP efflux transporter function. Further studies are needed to explore the functionality of GLUT1 and to what extent paracellular and transcellular transport is functional in the CS79-derived BECs.
Others have suggested that hiPSC-BECs in this protocol may not express VE-Cadherin expression and may represent an intermediate phenotype between neuroepithelium and brain endothelium (Lu et al., 2021). Here we demonstrate robust VE-Cadherin expression following this differentiation protocol suggesting an endothelial identity, however PECAM-1 expression was less convincing. Future work could expand on the differentiation by the inclusion of ETS transcription factors or small molecule cocktails that have been demonstrated to increase endothelial identity in this differentiation model (Porkoláb et al., 2023).
Overall, the CS79iBRCA-n2 derived BECs recapitulate some BBB-like properties such as high TEER, expression of tight junctions Claudin-5, ZO-1, and Occludin, and functional efflux transporters BCRP and P-gp.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
NA: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review and editing. KB: Data curation, Investigation, Methodology, Writing – original draft. AM: Writing – original draft, Writing – review and editing. LM: Data curation, Writing – original draft. CV: Writing – original draft. SR: Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review and editing. BK: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by research funding from the Breast Cancer Research Foundation of Alabama (BK, SR), The University of Alabama Carolyn Lawless and Janice Elizabeth Innes Research Award in Cell and Molecular Biology (NA), and The University of Alabama ASSURE Awards (AM, LM).
Acknowledgments
We thank Cedars-Sinai Biomanufacturing Center for their assistance obtaining the CS79iBRCA-n2 cell line.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbott, N. J., Patabendige, A. A. K., Dolman, D. E. M., Yusof, S. R., and Begley, D. J. (2010). Structure and function of the blood-brain barrier. Neurobiol. Dis. 37 (1), 13–25. doi:10.1016/j.nbd.2009.07.030
Allen, J. D., van Loevezijn, A., Lakhai, J. M., van der Valk, M., van Tellingen, O., Reid, G., et al. (2002). Potent and specific inhibition of the breast cancer resistance protein multidrug transporter in vitro and in mouse intestine by a novel analogue of fumitremorgin C. Mol. Cancer Ther. 1 (6), 417–425.
Andjelkovic, A. V., Stamatovic, S. M., Phillips, C. M., Martinez-Revollar, G., and Keep, R. F. (2020). Modeling blood–brain barrier pathology in cerebrovascular disease in vitro: current and future paradigms. Fluids Barriers CNS 17 (1), 44. doi:10.1186/s12987-020-00202-7
Aragón-González, A., Shaw, A. C., Kok, J. R., Roussel, F. S., Santos Souza, C. D., Granger, S. M., et al. (2024). C9ORF72 patient-derived endothelial cells drive blood-brain barrier disruption and contribute to neurotoxicity. Fluids Barriers CNS 21 (1), 34. doi:10.1186/s12987-024-00528-6
Arshad, F., Wang, L., Sy, C., Avraham, S., and Avraham, H. K. (2011). Blood-brain barrier integrity and breast cancer metastasis to the brain. Pathology Res. Int. 2011, 920509–920512. doi:10.4061/2011/920509
Ben-Zion Berliner, M., Yust-Katz, S., Lavie, I., Goldberg, Y., Kedar, I., and Yerushalmi, R. (2024). Central nervous system metastases in breast cancer patients with germline BRCA pathogenic variants compared to non-carriers: a matched-pair analysis. BMC Cancer 24 (1), 219. doi:10.1186/s12885-024-11975-7
Boulton, S. J. (2006). Cellular functions of the BRCA tumour-suppressor proteins. Biochem. Soc. Trans. 34 (5), 633–645. doi:10.1042/BST0340633
Curry, E. L., Moad, M., Robson, C. N., and Heer, R. (2015). Using induced pluripotent stem cells as a tool for modelling carcinogenesis. World J. Stem Cells 7 (2), 461–469. doi:10.4252/wjsc.v7.i2.461
Engdahl, E., Van Schijndel, M. D. M., Voulgaris, D., Di Criscio, M., Ramsbottom, K. A., Rigden, D. J., et al. (2021). Bisphenol A Inhibits the transporter function of the blood-brain barrier by directly interacting with the ABC transporter breast cancer resistance protein (BCRP). Int. J. Mol. Sci. 22 (11), 5534. doi:10.3390/ijms22115534
Espinal, E. R., Sharp, S. J., and Kim, B. J. (2022). Induced pluripotent stem cell (iPSC)-Derived endothelial cells to study bacterial-brain endothelial cell interactions. Methods Mol. Biol. 2492, 73–101. doi:10.1007/978-1-0716-2289-6_4
Gastfriend, B. D., Palecek, S. P., and Shusta, E. V. (2018). Modeling the blood-brain barrier: beyond the endothelial cells. Curr. Opin. Biomed. Eng. 5, 6–12. doi:10.1016/j.cobme.2017.11.002
Gomes, S. F. M., Westermann, A. J., Sauerwein, T., Hertlein, T., Forstner, K. U., Ohlsen, K., et al. (2019). Induced pluripotent stem cell-derived brain endothelial cells as a cellular model to study Neisseria meningitidis infection. Front. Microbiol. 10, 1181. doi:10.3389/fmicb.2019.01181
Gudmundsdottir, K., and Ashworth, A. (2006). The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene 25 (43), 5864–5874. doi:10.1038/sj.onc.1209874
He, Y., Yao, Y., Tsirka, S. E., and Cao, Y. (2014). Cell-culture models of the blood–brain barrier. Stroke 45 (8), 2514–2526. doi:10.1161/STROKEAHA.114.005427
Helms, H. C., Abbott, N. J., Burek, M., Cecchelli, R., Couraud, P. O., Deli, M. A., et al. (2016). In vitro models of the blood-brain barrier: an overview of commonly used brain endothelial cell culture models and guidelines for their use. J. Cereb. Blood Flow Metab. 36 (5), 862–890. doi:10.1177/0271678X16630991
Hersh, D. S., Wadajkar, A S., Roberts, N. B., Perez, G. J., Connolly, N. P., Frenkel, V., et al. (2016). Evolving drug delivery strategies to overcome the blood brain barrier. Curr. Pharm. Des. 22 (9), 1177–1193. doi:10.2174/1381612822666151221150733
Hollmann, E. K., Bailey, A. K., Potharazu, A. V., Neely, M. D., Bowman, A. B., and Lippmann, E. S. (2017). Accelerated differentiation of human induced pluripotent stem cells to blood–brain barrier endothelial cells. Fluids Barriers CNS 14 (1), 9. doi:10.1186/s12987-017-0059-0
Kim, B. J., McDonagh, M. A., Deng, L., Gastfriend, B. D., Schubert-Unkmeir, A., Doran, K. S., et al. (2019). Streptococcus agalactiae disrupts P-glycoprotein function in brain endothelial cells. Fluids Barriers CNS 16 (1), 26. doi:10.1186/s12987-019-0146-5
Knopp, R. C., Banks, W. A., and Erickson, M. A. (2022). Physical associations of microglia and the vascular blood-brain barrier and their importance in development, health, and disease. Curr. Opin. Neurobiol. 77, 102648. doi:10.1016/j.conb.2022.102648
Lim, R. G., Quan, C., Reyes-Ortiz, A. M., Lutz, S. E., Kedaigle, A. J., Gipson, T. A., et al. (2017). Huntington’s disease iPSC-derived brain microvascular endothelial cells reveal WNT-mediated angiogenic and blood-brain barrier deficits. Cell Rep. 19 (7), 1365–1377. doi:10.1016/j.celrep.2017.04.021
Lippmann, E. S., Al-Ahmad, A., Azarin, S. M., Palecek, S. P., and Shusta, E. V. (2014). A retinoic acid-enhanced, multicellular human blood-brain barrier model derived from stem cell sources. Sci. Rep. 4 (1), 4160. doi:10.1038/srep04160
Lippmann, E. S., Azarin, S. M., Kay, J. E., Nessler, R. A., Wilson, H. K., Al-Ahmad, A., et al. (2012). Derivation of blood-brain barrier endothelial cells from human pluripotent stem cells. Nat. Biotechnol. 30 (8), 783–791. doi:10.1038/nbt.2247
Lu, T. M., Houghton, S., Magdeldin, T., Durán, J. G. B., Minotti, A. P., Snead, A., et al. (2021). Pluripotent stem cell-derived epithelium misidentified as brain microvascular endothelium requires ETS factors to acquire vascular fate. Proc. Natl. Acad. Sci. U.S.A. 118 (8), e2016950118. doi:10.1073/pnas.2016950118
Mai, P. L., Chatterjee, N., Hartge, P., Tucker, M., Brody, L., Struewing, J. P., et al. (2009). Potential excess mortality in BRCA1/2 mutation carriers beyond breast, ovarian, prostate, and pancreatic cancers, and melanoma. PLoS One 4 (3), e4812. doi:10.1371/journal.pone.0004812
Mayer, U., Wagenaar, E., Dorobek, B., Beijnen, J. H., Borst, P., and Schinkel, A. H. (1997). Full blockade of intestinal P-glycoprotein and extensive inhibition of blood-brain barrier P-glycoprotein by oral treatment of mice with PSC833. J. Clin. Investig. 100 (10), 2430–2436. doi:10.1172/JCI119784
Nagai, H., and Kim, Y. H. (2017). Cancer prevention from the perspective of global cancer burden patterns. J. Thorac. Dis. 9 (3), 448–451. doi:10.21037/jtd.2017.02.75
Narkhede, A. A., Shevde, L. A., and Rao, S. S. (2017). Biomimetic strategies to recapitulate organ specific microenvironments for studying breast cancer metastasis. Int. J. Cancer 141 (6), 1091–1109. doi:10.1002/ijc.30748
Obermeier, B., Daneman, R., and Ransohoff, R. M. (2013). Development, maintenance and disruption of the blood-brain barrier. Nat. Med. 19 (12), 1584–1596. doi:10.1038/nm.3407
Ozgür, B., Puris, E., Brachner, A., Appelt-Menzel, A., Oerter, S., Balzer, V., et al. (2023). Characterization of an iPSC-based barrier model for blood-brain barrier investigations using the SBAD0201 stem cell line. Fluids Barriers CNS 20 (1), 96. doi:10.1186/s12987-023-00501-9
Pardridge, W. M. (2007). Blood–brain barrier delivery. Drug Discov. Today 12 (1-2), 54–61. doi:10.1016/j.drudis.2006.10.013
Patabendige, A., and Janigro, D. (2023). The role of the blood–brain barrier during neurological disease and infection. Biochem. Soc. Trans. 51 (2), 613–626. doi:10.1042/BST20220830
Patel, R., Page, S., and Al-Ahmad, A. J. (2017). Isogenic blood-brain barrier models based on patient-derived stem cells display inter-individual differences in cell maturation and functionality. J. Neurochem. 142 (1), 74–88. doi:10.1111/jnc.14040
Paturi, D. K., Kwatra, D., Ananthula, H. K., Pal, D., and Mitra, A. K. (2010). Identification and functional characterization of breast cancer resistance protein in human bronchial epithelial cells (Calu-3). Int. J. Pharm. 384 (1-2), 32–38. doi:10.1016/j.ijpharm.2009.09.037
Porkoláb, G., Mészáros, M., Szecskó, A., Vigh, J. P., Walter, F. R., Figueiredo, R., et al. (2023). Synergistic induction of blood-brain barrier properties. Proc. Natl. Acad. Sci. U.S.A. 121, e2316006121. doi:10.1073/pnas.2316006121
Ratner, E., Bala, M., Louie-Gao, M., Aydin, E., Hazard, S., and Brastianos, P. K. (2019). Increased risk of brain metastases in ovarian cancer patients with BRCA mutations. Gynecol. Oncol. 153 (3), 568–573. doi:10.1016/j.ygyno.2019.03.004
Rubin, L. L., and Staddon, J. M. (1999). The cell biology of the blood-brain barrier. Annu. Rev. Neurosci. 22 (1), 11–28. doi:10.1146/annurev.neuro.22.1.11
Siegel, R. L., Giaquinto, A. N., and Jemal, A. (2024). Cancer statistics, 2024. CA A Cancer J. Clin. 74 (1), 12–49. doi:10.3322/caac.21820
Stebbins, M. J., Gastfriend, B. D., Canfield, S. G., Lee, M. S., Richards, D., Faubion, M. G., et al. (2019). Human pluripotent stem cell-derived brain pericyte-like cells induce blood-brain barrier properties. Sci. Adv. 5 (3), eaau7375. doi:10.1126/sciadv.aau7375
Stebbins, M. J., Wilson, H. K., Canfield, S. G., Qian, T., Palecek, S. P., and Shusta, E. V. (2016). Differentiation and characterization of human pluripotent stem cell-derived brain microvascular endothelial cells. Methods 101, 93–102. doi:10.1016/j.ymeth.2015.10.016
Sun, H., Hu, H., Liu, C., Sun, N., and Duan, C. (2021). Methods used for the measurement of blood-brain barrier integrity. Metab. Brain Dis. 36 (5), 723–735. doi:10.1007/s11011-021-00694-8
Sun, J., Ou, W., Han, D., Paganini-Hill, A., Fisher, M. J., and Sumbria, R. K. (2022). Comparative studies between the murine immortalized brain endothelial cell line (bEnd.3) and induced pluripotent stem cell-derived human brain endothelial cells for paracellular transport. PLoS One 17 (5), e0268860. doi:10.1371/journal.pone.0268860
Urich, E., Lazic, S. E., Molnos, J., Wells, I., and Freskgård, P.-O. (2012). Transcriptional profiling of human brain endothelial cells reveals key properties crucial for predictive in vitro blood-brain barrier models. PLoS One 7 (5), e38149. doi:10.1371/journal.pone.0038149
Vatine, G. D., Al-Ahmad, A., Barriga, B. K., Svendsen, S., Salim, A., Garcia, L., et al. (2017). Modeling psychomotor retardation using iPSCs from MCT8-deficient patients indicates a prominent role for the blood-brain barrier. Cell Stem Cell 20 (6), 831–843. doi:10.1016/j.stem.2017.04.002
Verscheijden, L. F. M., Koenderink, J. B., De Wildt, S. N., and Russel, F. G. M. (2021). Differences in P-glycoprotein activity in human and rodent blood–brain barrier assessed by mechanistic modelling. Arch. Toxicol. 95 (9), 3015–3029. doi:10.1007/s00204-021-03115-y
Wang, H., Naghavi, M., Allen, C., Barber, R. M., Bhutta, Z. A., Carter, A., et al. (2016). Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388 (10053), 1459–1544. doi:10.1016/S0140-6736(16)31012-1
Wilhelm, I., and Krizbai, I. A. (2014). In vitro models of the blood–brain barrier for the study of drug delivery to the brain. Mol. Pharm. 11 (7), 1949–1963. doi:10.1021/mp500046f
Keywords: blood-brain barrier, patient derived stem cells, BRCA1 mutation, brain endothelial cells, induced pluripotent stem cells, disease modeling
Citation: Alexander NG, Buchanan KA, Meyer AE, Mitterway LM, Vanderburgh CO, Rao SS and Kim BJ (2025) Using a brain-like endothelial cell differentiation to characterize the CS79iBRCA-n2 BRCA1 mutated patient derived stem cell line. Front. Cell Dev. Biol. 13:1516669. doi: 10.3389/fcell.2025.1516669
Received: 24 October 2024; Accepted: 11 April 2025;
Published: 30 April 2025.
Edited by:
Ivan Garcia-Bassets, San Diego State University, United StatesReviewed by:
Angels Almenar, University of California, San Diego, United StatesMoriah E. Katt, West Virginia University, United States
Copyright © 2025 Alexander, Buchanan, Meyer, Mitterway, Vanderburgh, Rao and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brandon J. Kim, QnJhbmRvbi5LaW1AVVREYWxsYXMuZWR1
 Natalie G. Alexander
Natalie G. Alexander Kylie A. Buchanan
Kylie A. Buchanan Alexandra E. Meyer
Alexandra E. Meyer Lauren M. Mitterway
Lauren M. Mitterway Caroline O. Vanderburgh
Caroline O. Vanderburgh Shreyas S. Rao
Shreyas S. Rao Brandon J. Kim
Brandon J. Kim